What is our mission?
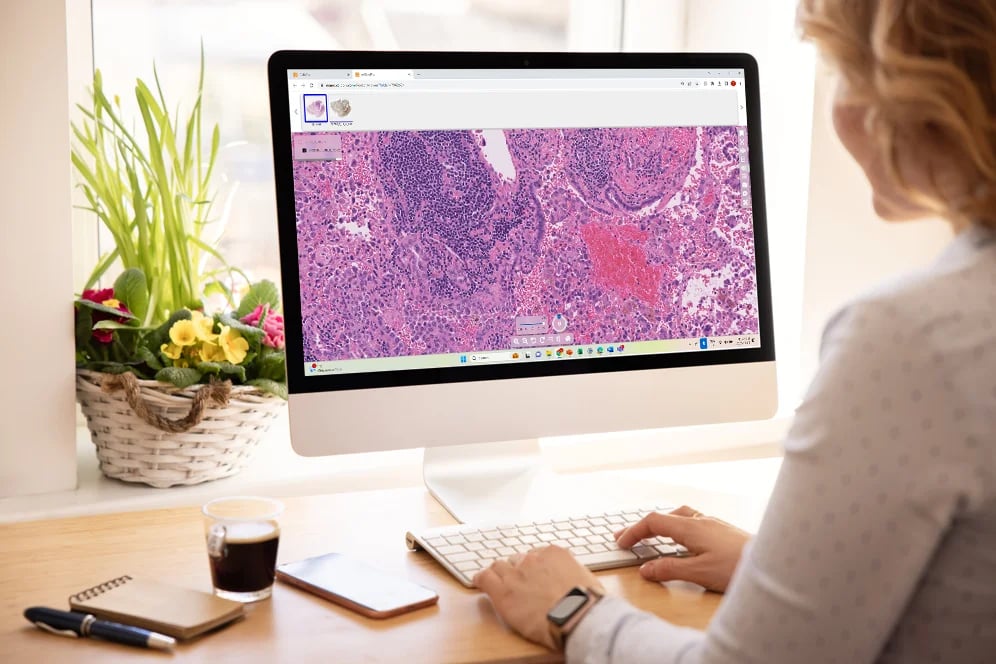
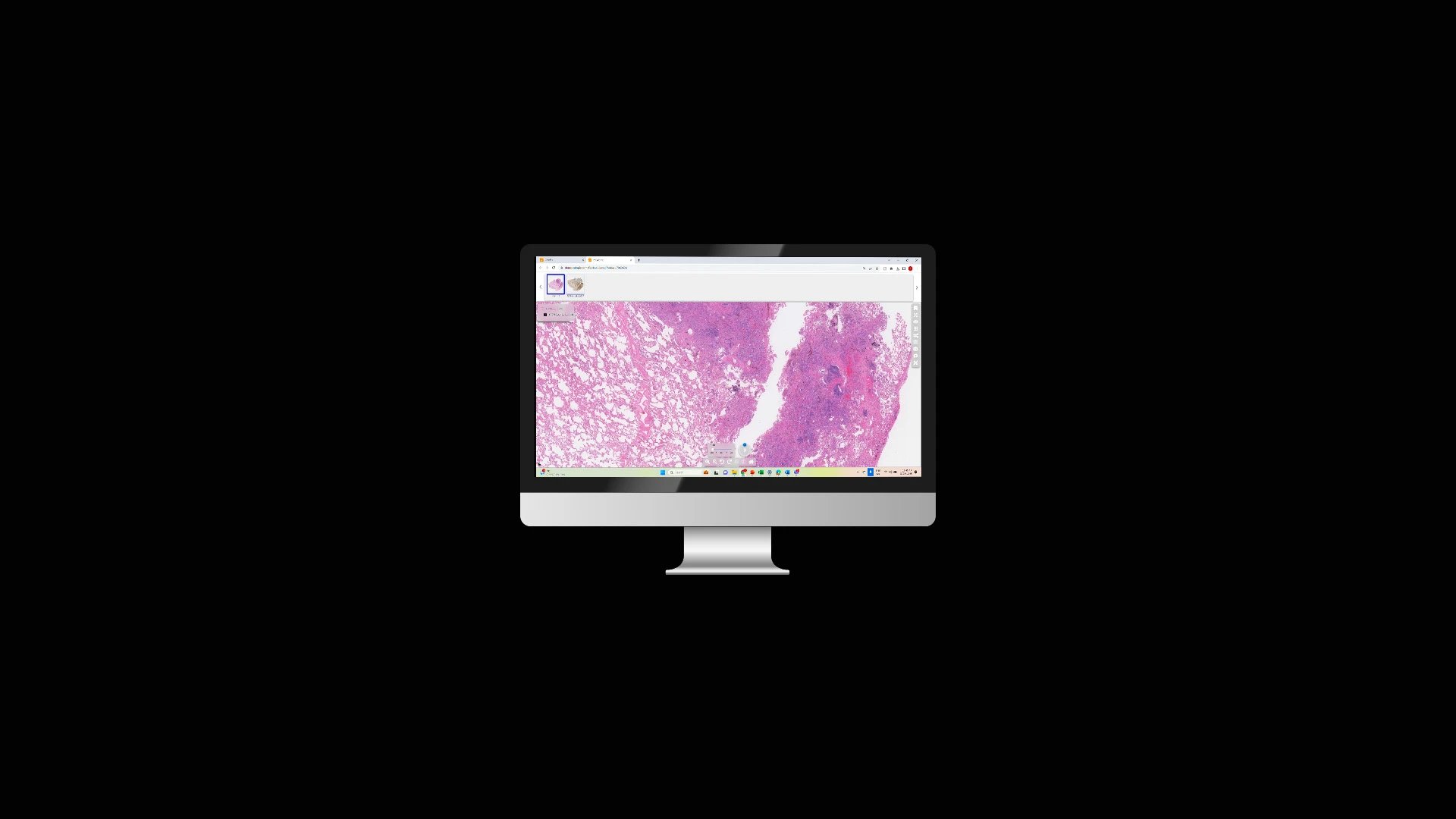
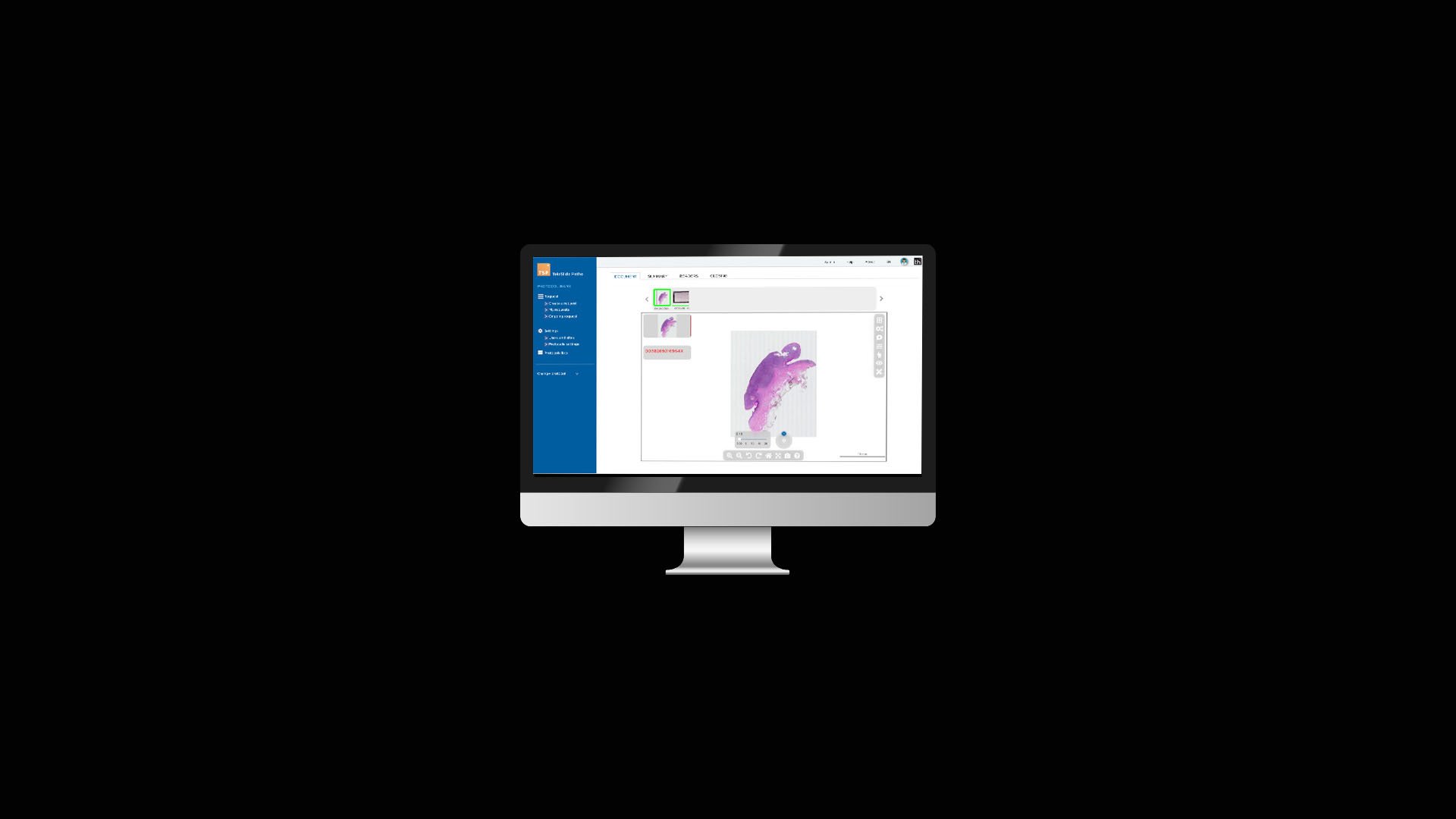
At Tribun Health, we're on a mission to modernize pathology diagnostics worldwide. Our innovative solutions, driven by artificial intelligence, propel the digital transformation in cancer diagnostics. By eliminating microscope dependencies, our software accelerates disease comprehension, progression tracking, and treatment identification.
In an era marked by dwindling medical resources and escalating specialization, quick access to expert diagnoses is paramount. Digitization in pathology labs emerges as the essential solution to address these evolving needs. Tribun Health stands ready to assist!




.png?width=256&height=256&name=customer-service(1).png)





.png?width=64&height=64&name=calendrier(1).png)
.png?width=64&height=64&name=communique-de-presse(1).png)
.png?width=64&height=64&name=livre(1).png)
.png?width=64&height=64&name=blog(2).png)